1. Protein Structure :
Central dogma
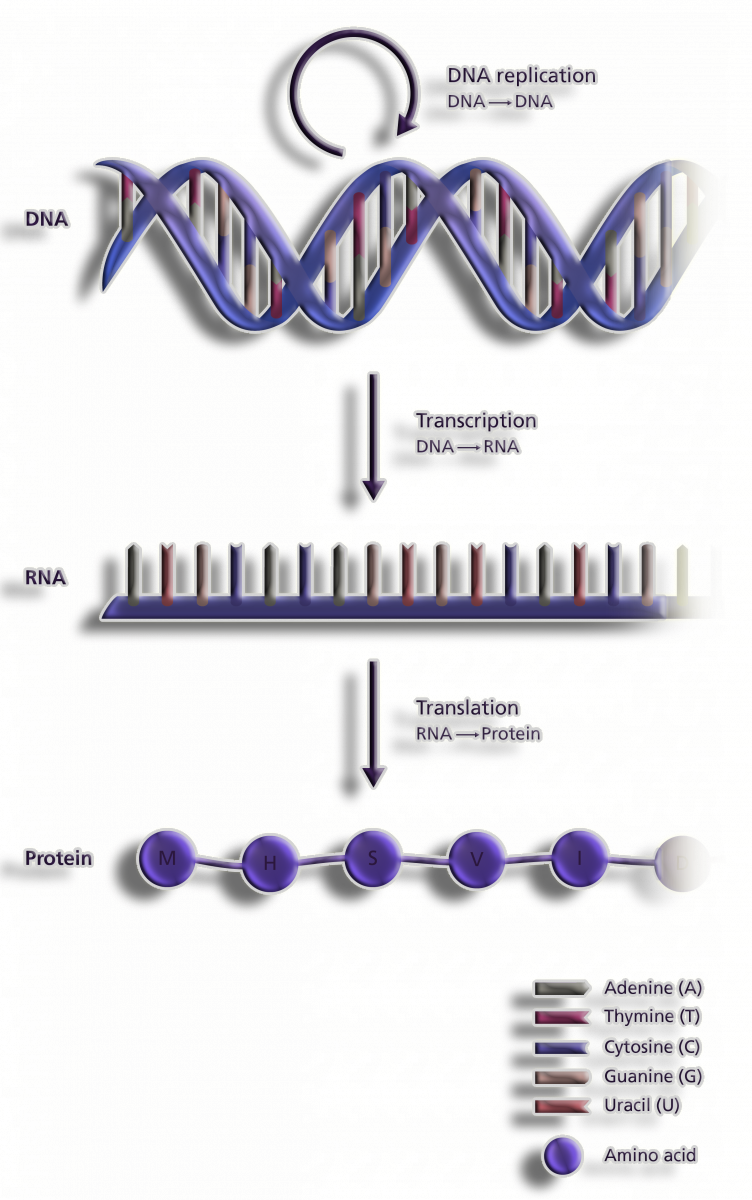
Figure 1. Central dogma of Life
Basic structure of Amino Acids
-
Its a linear polymer of amino acids forms proteinaceous structures.
-
The 3D structure of the protein is determined by the distinctive amino acid sequence.
-
The fundamental structure of a protein is its ordered, linear arrangement of amino acids.
Protein amino acid sequences can be used to differentiate them, according to Frederick Sanger (1952). Proteins with the exact same kind have unique sequences.
Definition of Central Dogma
A Central Dogma describes how DNA is converted to RNA and then into Proteins in Molecular Biology. A genomic process refers to the conversion of DNA information into a functional product.
Steps of the Central Dogma takes place in two tages:
1.Transcription
2.Translation
Transcription
The enzyme RNA Polymerase transfers information from one strand of DNA to another strand of RNA during transcription.
Three parts of the DNA strand are involved in this process: the promoter, the structural gene, and the terminator.
DNA strands that synthesize RNA are called template strands, and DNA strands that Code for RNA are called coding strands.
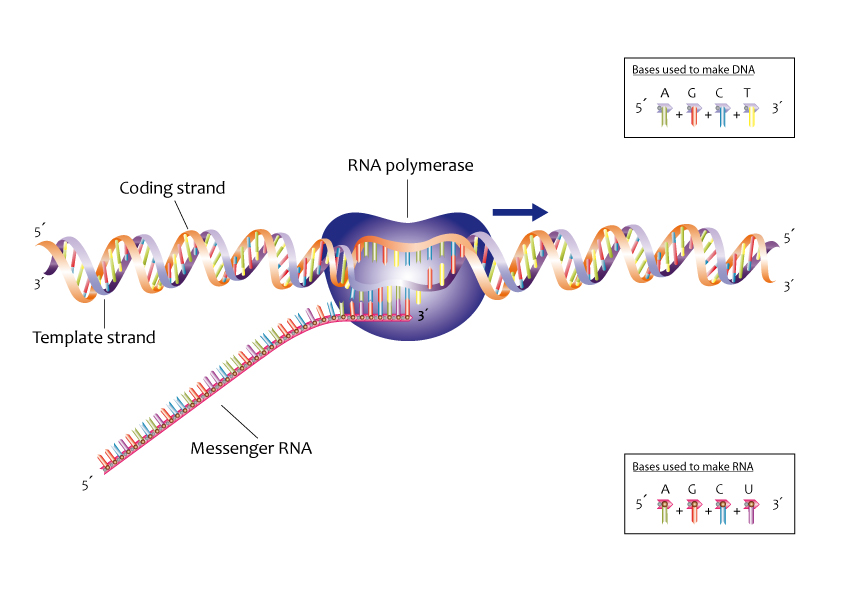
Figure 2. The process of creating an RNA copy of a gene's DNA sequence is known as transcription in the context of genomics. The DNA-encoded protein information for the gene is carried by this copy, known as messenger RNA (mRNA).
Translation
Proteins are encoded by RNA by a process called translation. Translation involves energy that comes from the charged tRNA Molecules.
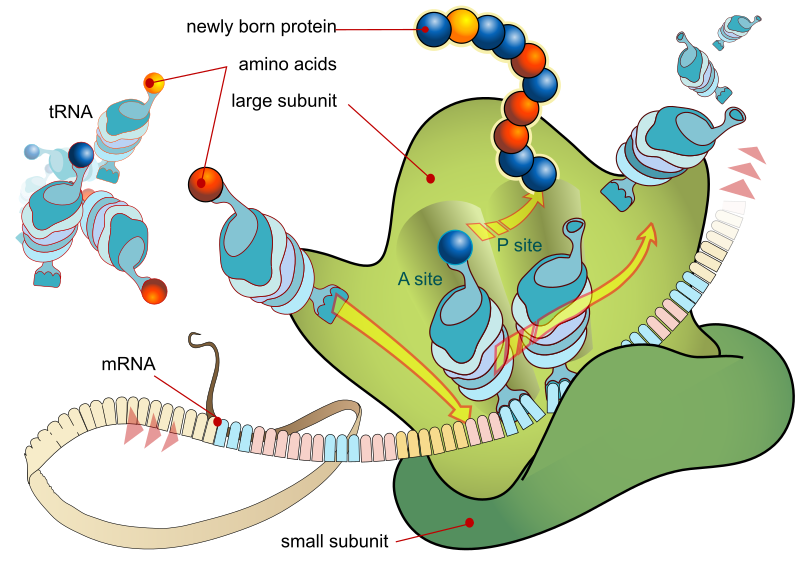
Figure 3. The process of producing proteins by ribosomes in the cytoplasm or endoplasmic reticulum is known as DNA translation.
The translation process is initiated by ribosomes which are made up of two subunits, one larger and one smaller.(larger subunit consists of two tRNA Molecules)
The mRNA enters the smaller subunit and is then held by the tRNA Molecules present in the larger subunit that are complementary to the codon.
Two amino acids are held together by two tRNA Molecules placed close together and a peptide bond is formed between them.
Three nucleotides and four nitrogenous bases collectively code for an amino acid, forming a triplet codon.there are 64 codons, of which three are stop codons (UAA, UAG and UGA) that end transcription and one is an initiator codon, i.e. AUG, which Codes for methionine.
Protein Hierarchy :
-
The primary structure of a protein is its amino acid sequence.
-
The secondary structure is the initial folding of the sequence (process of folding) into alpha helices and beta sheets.
-
The tertiary structure is a more complex folding of the protein upon itself.
-
The quarternary structure is the combination of two or more of the same protein.
-
And finally, the supramolecular structure is the combination of several different protein subunits.
It is this 3-D structure that determines the function of the protein, either for signaling, transport, catalysis, movement, structure, or regulation. AAs determine the uniqueness and seq of the protein and its folding nature and ultimately the function that it performs.

Figure 4. English biochemist Frederick Sanger twice received the Chemistry Nobel Prize.
Amino Acid Composition
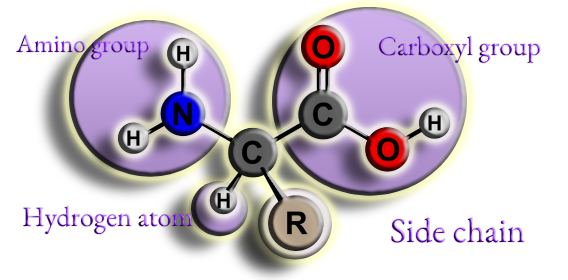
Figure 5. Skeleton structure of a common amino acid.
-
An amino group (NH2)
-
A carboxyl group (COOH)
-
An atom of hydrogen (H)
-
Side chain (or R group)
These groups are joined to an α-carbon in the middle.
A combination of 20 common amino acids is typically seen in proteins.

Figure 6. 20 common amino acids and their structures are listed above.
Amino acids can be classified into four general groups based on the properties of the "R" group in each amino acid. Amino acids can be :
-
Polar
-
Non-polar
-
Positively charged
-
Negatively charged
Polar amino acids have "R" groups that are hydrophilic, meaning that they seek contact with aqueous solutions.
Nonpolar amino acids are the opposite (hydrophobic) in that they avoid contact with liquid.
The nonpolar amino acids are hydrophobic, while the remaining groups are hydrophilic.
Basic, positively
charged amino acids
At physiological pH, the amino ends (NH4) of the radical groups of these
amino acids are unbalanced, and give the amino acids an overall positive
(basic) charge. These amino acids are also polar and therefore
hydrophilic.
Acidic, negatively
charged amino acids
At physiological pH, the carboxyl ends (COOH) of the radical groups of
these amino acids are unbalanced, and the amino acids have an
overall negative (acidic) charge. These amino acids are also polar and
therefore hydrophilic.
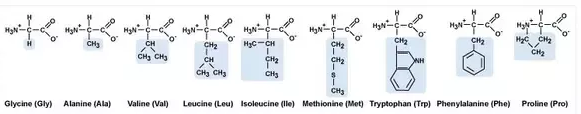
Nonpolar Amino Acids
Ala: Alanine Gly: Glycine Ile: Isoleucine Leu: Leucine
Met: Methionine Trp: Tryptophan Phe: Phenylalanine Pro: Proline
Val: Valine (GAIL MV PPT)
<----------------------------------------------------------------------------------------------------------->
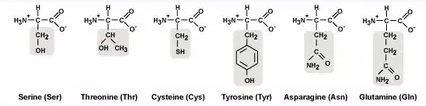
Polar Amino Acids
Cys: Cysteine Ser: Serine Thr: Threonine
Tyr: Tyrosine Asn: Asparagine
Gln: Glutamine (GAS CTT)
<----------------------------------------------------------------------------------------------------------->
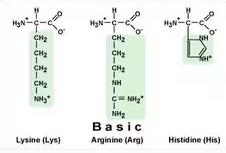
Polar Basic Amino Acids (Positively
Charged)
His: Histidine Lys: Lysine Arg: Arginine
(HAL)
<----------------------------------------------------------------------------------------------------------->
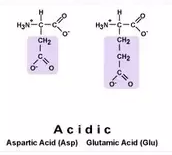
Polar Acidic Amino Acids (Negatively
Charged)
Asp: Aspartate Glu: Glutamate
(GA)
Of the 20 amino acids, 11 can be produced naturally.
These nonessential amino acids are alanine, arginine, asparagine, aspartate, cysteine, glutamate, glutamine, glycine, proline, serine, and tyrosine. Nonessential amino acids are synthesized from products or intermediates of crucial metabolic pathways.
A
stereocenter is always present if a
given carbon atom is connected to four different substituents.
L- and D-amino acids are the two
isomeric forms of amino acids which occur in nature.
L-amino acids are the form used by the cells to synthesize
proteins.
| L-Amino Acids |
D-Amino Acids |
| Amino group present
on the left side of fischer projection |
Amino group present
on the right side of fischer projection |
| Rotate plane
polarized light counterclockwise (Levorotation) |
Rotate plane
polarized light anticlockwise (dextrorotation) |
| R Notation |
S Notation |
| Used by cells to
produce proteins |
Occur in the cell
wall of bacteria |
L-Amino Acids
-
L-amino acids are the form of stereoisomers used by the cells to produce proteins.
-
They occur in all proteins produced by animals, plants, fungi, and bacteria.
-
These proteins have both structural and functional roles inside the cell. They serve as enzymes, which catalyze biochemical reactions, as well as hormones, which regulate the biological process in higher organisms.
D-Amino Acids
-
D-amino acids are the other form of stereoisomers that occur in nature.
-
The amine group of these amino acids occurs in the right side in the Fisher projection. cells do not incorporate D-amino acids into proteins.
-
But, some proteins are produced by enzyme posttranslational modifications in cone snails.
-
On the other hand, some D-amino acids also occur in the peptidoglycan cell walls of bacteria. In addition, D-serine serves as a neurotransmitter in the brain.
L- and D-amino acids are two possible orientations of a particular amino acid in nature.
They are the mirror image of each other.
Also, they can be considered as the isomeric forms, stereoisomers or enantiomers.
However, the simplest amino acid, glycine, does not have stereoisomers. Because it lacks a chiral carbon atom or a tetrahedral carbon atom, glycine is optically inactive. Glycine lacks optical activity because it lacks an alkyl group and instead possesses a hydrogen atom.
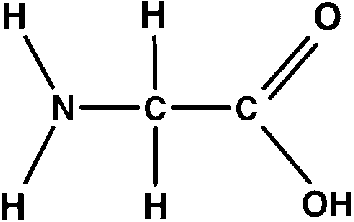
Both contain a carboxylic acid group, an amine group, a carbon chain, and a hydrogen atom bound to the central carbon atom of the amino acid. Further, this central carbon atom is called the alpha carbon or the chiral carbon.
Difference Between L and D Amino Acids
Definition
L-amino acid refers to a stereoisomer of a particular amino acid whose amino group is on the left side in its Fisher projection while D-amino acid refers to the other stereoisomer of the amino acid whose amino group is on the right side in its Fisher projection. This explains the basic difference between L and D amino acids.
Rotation of the Plane-Polarized Light
Also, while L-amino acids can rotate plane-polarized light counterclockwise in a process called levorotation, D-amino acids can rotate plane-polarized light clockwise in a process called dextrorotation.
R/S Notation
Another difference between L and D amino acids is that L-amino acids have been superseded by R notation while the D-amino acids have been superseded by S notation.
Importance
Moreover, L-amino acids are used by the cell
to produce proteins while D-amino
acids occur in the cell wall of bacteria.
Conclusion
L-amino acids are a form of stereoisomers whose amine group occurs in
the left side of the Fisher projection. On the other hand, D-amino acids
are the other form of stereoisomers whose amine group occurs on the
right side of the Fisher projection. L-amino acids are a form of
stereoisomer which is abundant in proteins. This is the difference
between L and D amino acids.
Color schemas are commonly used in case of amino acids 3D structure representation.
Color schemas for representation in biomolecules are seen commonly, some are listed below :
Carbon Grey
Hydrogen White
Oxygen Red
Nitrogen Blue
Sulphur Yellow
Phosphorus Orange
The genetic code refers to the entire set of interactions between codons and amino acids (or stop signals). In a codon chart (or codon table), where codons are translated to amino acids, the genetic code is frequently condensed.

Figure 7. Marshall W. Nirenberg, Robert W. Holley, and Har Gobind Khorana all shared the 1968 Nobel Prize for Physiology or Medicine for their work demonstrating the sequence of nucleotides in nucleic acids, which contain cellular genetic information and regulate protein synthesis.
Aromatic amino acids are amino acids that have an aromatic ring in the side-chain.Four of the 21 standard amino acids have aromatic rings in their side-chains.
The diagram below shows the structures of phenylalanine, tyrosine, tryptophan, and histidine.
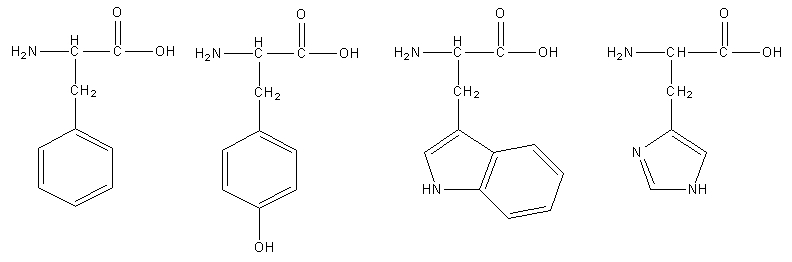
Figure 8. 2D structures of phenylalanine, tyrosine, tryptophan, and histidine.
Polar atoms that are either a component of or linked to the aromatic systems further distinguish tryptophan from other amino acids.
-
Amino acids are crystalline, colorless solids.
-
The melting point of all amino acids is higher than 200 °C.
-
They are somewhat soluble in alcohol and soluble in water, when heated to high temperatures, they breakdown.
-
Except for glycine, all amino acids have optical activity.
Zwitterionic property
-
A zwitterion is a molecule with functional groups, of which at least one has a positive and one has a negative electrical charge.
-
The net charge of the entire molecule is zero. Amino acids are the best-known examples of zwitterions.
-
They contain an amine group (basic) and a carboxylic group (acidic).
-
The -NH2 group is the stronger base, and so it picks up H+ from the -COOH group to leave a zwitterion. The (neutral) zwitterion is the usual form of amino acids that exist in the solution.
Amphoteric property
-
Amino acids are amphoteric in nature that is they act as both acids and base due to the two amine and carboxylic groups present.
Disulphide Bond
A disulfide bond is a single covalent connection created when thiol (SH) units are coupled together. They are created in proteins in the space between cysteine residues' thiol groups. The linkage is also known as a "disulfide bridge" or a "S-S bond."
.png)
Figure 9. Di-sulphide bond formation.
Some proteins' folding and stability depend heavily on disulfide bonds. A protein's folded state is stabilized by the disulfide bond in a number of ways.
-
It binds two distinct regions of the protein together, favoring the folded structure.
-
Local hydrophobic residues may condense around the disulfide bond and onto one another through hydrophobic interactions, forming the nucleus of a hydrophobic core of the folded protein.
An essential part of the secondary and tertiary structure of proteins is the disulfide bridges that are produced between the thiol groups in two cysteine residues.
A protein's folded state is stabilized by the disulfide bond in a number of ways.
-
It binds together two sections of the protein, favoring the folded structure. In other words, the disulfide bond reduces the entropy of the protein's unfolded state, which destabilizes it.
-
Local hydrophobic residues may condense around the disulfide link and onto one another through hydrophobic interactions, forming the nucleus of a hydrophobic core of the folded protein.
-
In relation to points 1 and 2, a disulfide bond joins two protein chain segments, raising the effective local concentration of protein residues while lowering the effective local concentration of water molecules.
Peptide Bonds
They are chains of amino acids, which are linked by peptide bonds.
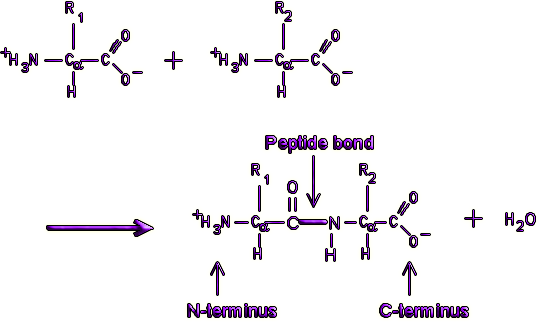
Figure 10. Peptide bond formation.
Every end of polypeptide is called amino terminal or N terminal, which has a free amino group. The other end of polypeptide has a free carboxyl group called C terminal or carboxyl terminal.
Polypeptides play an important part of proteins in the cells. Proteins consist of one or more molecules of polypeptides. Proteins are important as they help the cells in various ways.
Polypeptides make proteins by bonding together various amino acids. Two or more polypeptides bond and fold into a specific shape to form a particular protein.
.png)
Figure 11. More complicated polymer chains can be created by linking together single amino acid monomers.
The other name for polypeptide is an amino acid polymer. They are chains of monomers and subunits linked together by a chemical bond. A single chain of a polypeptide is called simple protein.
Polypeptides have various important functions and their function is dependent on their size and amino acid sequence.
A chain of between twenty and fifty amino acids joined together by covalent peptide bonds is referred to as a polypeptide.
The building blocks of life are single amino acids, which can be combined inside of cells to create oligopeptides, polypeptides, and proteins. This takes place during a procedure known as protein synthesis.
The twenty-one essential and non-essential amino acids can be arranged in a variety of ways to create a huge variety of functional peptide-based compounds.
Oligopeptides are linear chains of between two and twenty amino acids.
A dipeptide – the result of two bonded amino acids – is also an oligopeptide.
Linear chains of between twenty and fifty amino acids are called polypeptides.
These can form primary or secondary structures. Larger peptides are categorized according to their structure. Proteins contain at least fifty amino acids and often wrap into distinct shapes. They are grouped into primary, secondary, tertiary, and quaternary structures.
-
A simple peptide chain's amino acid sequence is referred regarded as having a primary structure.
.png)
Figure 12. The sequence of amino acids joined together to form a polypeptide chain is referred to as a protein's fundamental structure.
-
Secondary structures, which are frequently alpha-helix, beta-strand, or beta-sheet, use hydrogen bonds to construct spirals, coils, or sheets from a single polypeptide chain.
.png)
Figure 13. The term "secondary structure" describes the regular, local structure of the protein backbone that is stabilized by amide group hydrogen bonds that occur both within and outside of individual molecules.
-
Tertiary structure similarly involves a single polypeptide chain, but it folds the secondary structure further to create a globule-like molecule that is, for instance, held in place by salt bridges and hydrogen bonds.
.png)
Figure 14. A protein's overall three-dimensional arrangement of its polypeptide chain in space is referred to as its tertiary structure. It is primarily maintained by outer polar hydrophilic hydrogen and ionic bond contacts, and inside hydrophobic interactions between nonpolar amino acid side chains.
-
Two or more polypeptide chains combine to form a protein with a single function in quaternary structures. The oxygen-transporting molecule in our blood, hemoglobin, is a quaternary protein composed of two bonded pairs of polypeptide chains.
.png)
Figure 15. An essential feature of proteins that is directly linked to their function is their quaternary structure. Oligomeric proteins are proteins that have quaternary structure. Numerous biological processes, including metabolism, signal transduction, and chromosome replication, are aided by oligomeric proteins.
Peptide Bond formation
A ribosome uses peptide bonds to join two amino acids together.
The carboxyl group of one amino acid and the amino group of the other form covalent chemical bonds. When these two residues bond, they release a single molecule of water — a dehydration synthesis reaction.
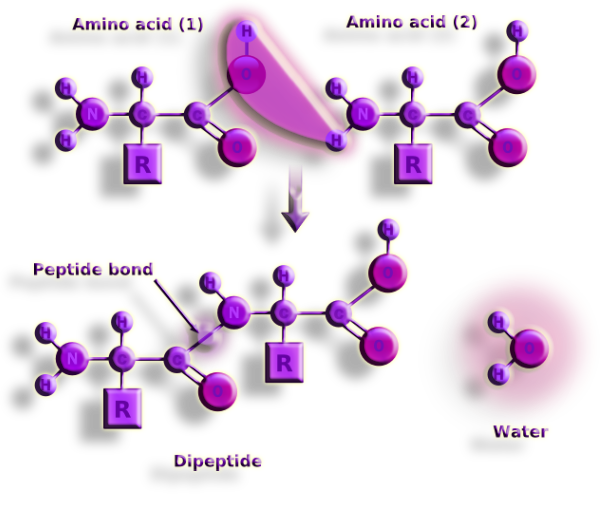
Figure 16. Dehydration synthesis is the technique used to create peptide bonds. Dehydration synthesis, commonly known as polymerization, is an anabolic procedure that involves the removal of a water molecule from connect monomers to produce a polymer.
To create H20, the amino group loses a positively charged hydrogen ion and the carboxyl group loses its negatively charged hydroxyl ion. This process is also known as a condensation reaction because the connected amino acids dehydrate and water is created.
This loss of ions from the newly bonded amino acid molecules explains why many textbooks claim that amino acid residues are the building blocks of polypeptides and proteins.
The ribosome assists in bringing the carboxyl group of one amino acid closer to the amino group of the following amino acid in order to form a peptide bond. Adenosine triphosphate is needed as energy for the creation of a peptide bond (ATP).
The dehydration synthesis step can be reversed by adding water, however this breaks peptide bonds much more slowly than the former procedure. This is owing to the additional double bond found in amino acid residues.
In the body, enzymes speed up this reaction. The positive charge of the carbon atom in the carboxyl is degraded by the addition of water, causing the bond to weaken.
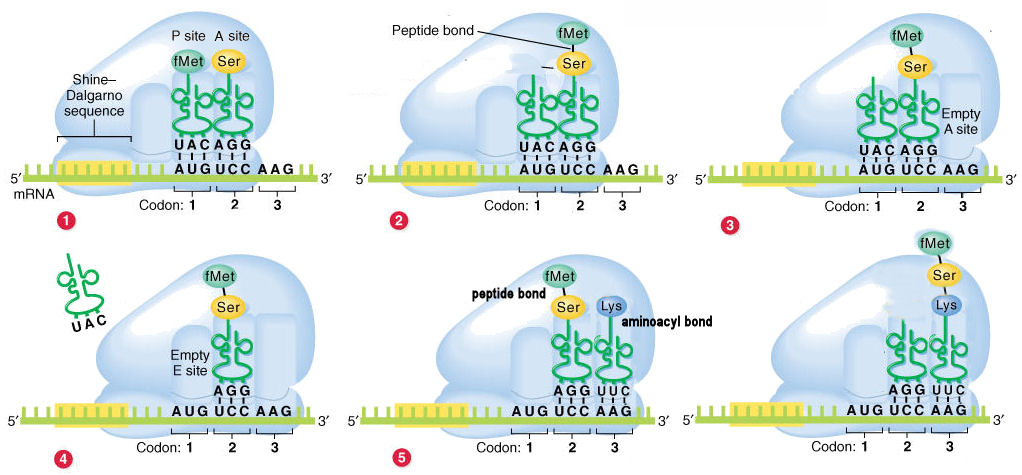
Figure 17. The condensation process between the carboxyl group of one amino acid and the amino group of another produces peptides in a chemical reaction.
The diagram presents the messenger RNA
(mRNA) molecule as "fixed", with the 5'3' direction oriented left to
right". The Ribosome "moves" along the
message from left to right, thus the mRNA is read 5'3'
direction [or, the mRNA passes through the Ribosome in the 5'3'
direction]. Note that the Ribosome has already encountered the
Shine-Delgarno promoter sequence in the mRNA.
Given this orientation, the diagram therefore shows the
left-to-right order of the Exit, Peptidyl, &
Aminoacyl sites as EPA. However, the functional order is APE,
since successive tRNAs enter through the A site, pass to the P site, and
exit from the E site.
(1) The fMet-tRNA initiates the process is
present at
the P site. The next charged tRNA enter the A site.
(2) The fMet is cleaved from the tRNA
in the P site, and linked by a peptide bond
to the amino acid in the A site.
(3) The tRNA in the P sites moves to the E
site, and the tRNA in the A site with the di-peptide moves to the
P site.
(4) The uncharged tRNA is released from the E
site.
(5) A new charged tRNA enters the A site.
At this point, the P site contains a peptidyl
bond, and the A site an aminoacyl bond.
(6) The di-peptide is cleaved from the tRNA
in the P site, and linked by a peptide bond to the amino acid in
the A site (as in step 2 above). This produces a tri-peptide in the A
site.
The cycle continues in this manner. The tRNAs in the P and A sites shift
to the E and P sites, the uncharged tRNA in the E site drops out, a new
tRNA with the amino acid for the next codon enters the A sites, and so
on.
Transfer of the amino acid from the first tRNA to the second tRNA may at
first seem counter-intuitive. This however ensures that the original
amino terminus of the first amino acid always remains unmodified: thus
the polypeptide "grows" in the amino carboxy (NC).
The characteristics of peptide bond:
-
Relevant implications for protein three-dimensional structure.
-
Character with some double entendre (resonance structures).
-
The peptide link is rigid and planar.
.png)
Figure 18. The peptide group's overall structure is rigid and flat. The resonance of the amide is what gives peptide bonds their stability. The double bond of the carbonyl group interacts with the C-N bond's electrons to generate a resonance structure.
Resonance structures of the peptide have an average C-N bond lengths:
1.27 Å (C=N bond)
1.49 Å (C—N bond)
1.32 Å (peptide bond)
(1 Å = 1xl0^-10 m)
Torsion angles
A torsion angle is formed by four atoms
consecutively bonded.
Not to be confused with systems where three atoms are bonded
to a central atom. (out of plane bending)
.png)
Figure 19. Torsion angles are depicted as Phi,Psi and Omega angles.
The polypeptide chain only has rotational freedom about the bonds formed by the α-carbons, Phi (Φ) and Psi (Ψ) angles.
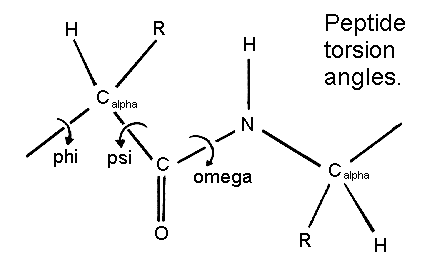
.png)
Figure 20. The conformation of the backbone can be described by two dihedral angles per residue, because the backbone residing between two juxtaposing Cα atoms are all in a single plane. These angles are called φ (phi) which involves the backbone atoms C-N-Cα-C, and ψ (psi) which involves the backbone atoms N-Cα-C-N.
A polypeptide's full structural space is mapped using a Ramachandran plot, which plots ('Phi' vs 'Psi' angles) and designates "permitted" and "disallowed" conformations. established by G.N. Ramachandran in the late 1960s as a result of research on combinations of sterically permitted "phi" and "psi" angles.
.jpg)
.png)
Figure 21. A Ramachandran plot in biochemistry was first created in 1963 is a method to show energetically permitted ranges for backbone dihedral angles versus of amino acid residues in protein structure was developed by G.N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan.
Ramachandran plot exceptions : Glycine and Proline
-
Glycine only possesses a hydrogen as a "side chain," as opposed to every other amino acid. It is less constrained due to its smaller van der Waals radius. The only amino group without a chiral center is glycine.
-
Proline, on the contrary, has a side chain that is a five-membered ring. As a result, it is significantly more constrained than the other amino acids and only permits a small quantity of phi and psi. The only amino acid with a 5-membered aliphatic ring system is proline.
.jpg)
.jpg)
Figure 22.
Since
Glycine has
a hydrogen atom as its R group, glycine is an exception to the
Ramachandran plot, while in
Proline has
its side chain which cyclizes onto the backbone forming a ring structure
in which a secondary amino group replaces the primary amino group.
Secondary structure of protein
Secondary structure refers to the local conformation of the polypeptide chain that exists independently from the remainder of the protein.
3D-Figure 1. PDB structure of 1FSV is a beta beta alpha motif with sequence length of 28 (CLICK me for more info!!).
The restrictions that the peptide bond and hydrogen bonding place on the main structure determine the secondary structure.
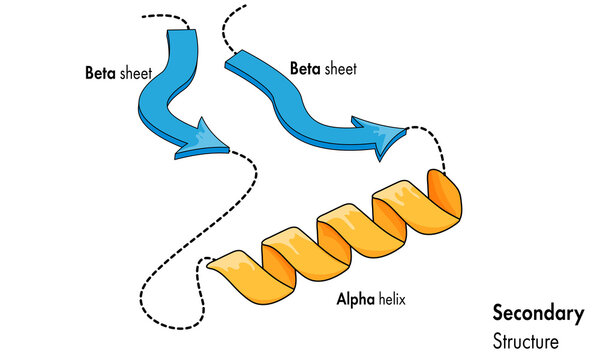
Figure 23. Beta-Pleated sheets and alpha-Helixes are protein secondary structure, created by hydrogen bonds between the amino H and carbonyl O of different amino acids.
The two main categories of secondary structure are Beta-sheets and alpha-helices. On the Ramachandran plot, the alpha helices and beta sheets have phi and psi angles that fall within the two largest permitted conformational areas.
Alpha helix :
The polypeptide backbone is bent to produce a
regular coil shape, which is the shape of a helix.
Right-handed helixes have coils that face to the right.
Since steric constraints prevent left-handed helices from
forming, almost all helices found in proteins are right-handed.
.png)
Figure 24. A secondary structure, or the description of how the primary chain of a protein is structured in space, is an alpha helix. It is a repeated regular secondary structure, meaning all of the residues have a similar conformation and hydrogen bonding, and it can be any length (much like the beta strand).
The alpha-helices are by far the most common right-handed helices.
3.6 residues make up an alpha-helix, which has a period of one turn of the backbone coil.
The carbonyl oxygen of each residue and the amide proton of the residue four positions ahead of it in the helix form hydrogen bonds, which stabilize the alpha-helix.
Backbone hydrogen bonds in every configuration are satisfied.
3D-Figure 2. PDB structure of 1COS is a triple-stranded alpha-helical bundle with sequence length of 31 (CLICK me for more info!!).
Various other helices
Three residues are
present on each turn of the 3-10 helix,
and hydrogen three locations of bonds exist
between each residue and the residue.
ahead.
.png)
Figure 25. The α-helix can be described as a 3.613 helix, since the i + 4 spacing adds three more atoms to the H-bonded loop compared to the tighter 310 helix, and on average, 3.6 amino acids are involved in one ring of α-helix.
The extremely uncommon n helix has 4.4 residues per turn, with each residue forms hydrogen bonds with residue five has only been observed at the ends of or-helices and positions ahead.
Beta sheets
-
Beta-sheets are created through hydrogen bonds between contiguous chains of polypeptides.
-
Polypeptide chain fragments taking part in the sheet include called beta-strands.
-
beta-strands are a prolonged conformity with the chain of polypeptides, where phi and psi angles are turned about 180 degrees with regard to one another.
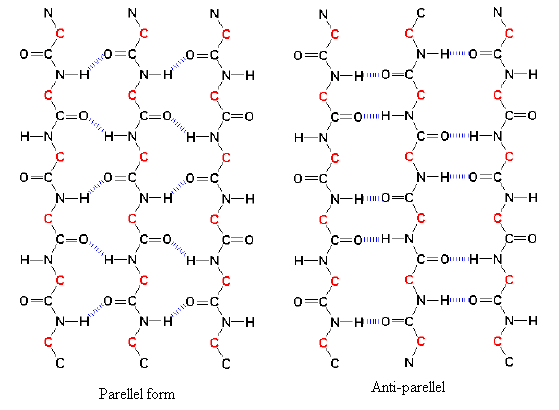
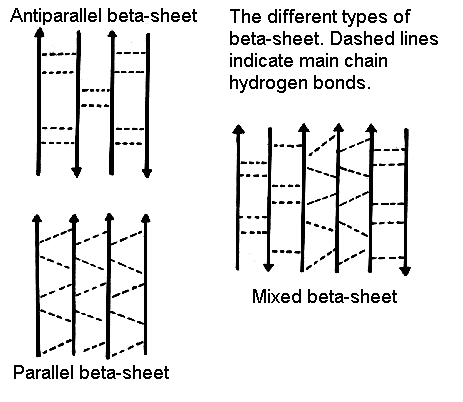
Figure 26. Beta sheets come in two varieties: parallel and anti-parallel sheets. Chains of polypeptides called parallel beta sheets run in the same direction. Chains of polypeptides known as anti-parallel beta sheets flow counterclockwise from one another.
There are two variations of the beta-sheet which include Parallel and antiparallel with both being feasible.
The sheets of parallel strands are set up in the same direction of their N-terminal amino acids and carboxy (C) terminal ends.
When sheets are antiparallel, the strands switch between amino and carboxy terminal ends, such that a specific strand engages with strands running the other way orientation.
.png)
Figure 27.
The geometry of each
individual amino acid molecule compels the hydrogen bonds to form at an
angle, lengthening and weakening them, making
the parallel arrangement less stable.
Due to the more ideal hydrogen bonding structure,
antiparallel beta sheets are known to be
slightly more stable than parallel beta sheets.
B-sheets can also develop; further In very few cases, when blended, parallel and antiparallel sections as mixed.
Beta-strands must interact with one another in order to form a sheet. The backbone of an amino acid must be turned.

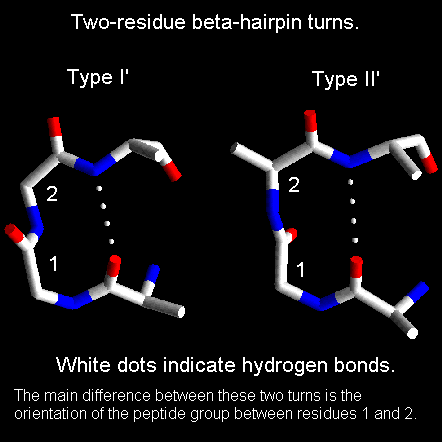
Figure 28. Two -strands connected by a turn or a brief loop form a tiny protein structural motif known as a "hairpin" when they fold to establish hydrogen bonds with one another. The interactions between nonlocal amino acids in -hairpins are different from those in helices.
The B-hairpin turn is the easiest and narrowest turn (also called type I and type ll turns, depending on the conformation of the backbone).
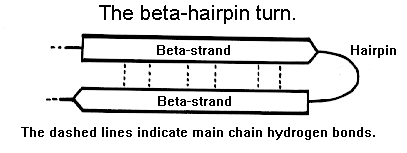
Figure 29. One of the simplest supersecondary structures, beta-hairpins are present in many globular proteins. Between beta-strands that are hydrogen linked in an antiparallel fashion, they appear as brief loop regions.
The most prevalent type of turn, occurring often between opposing strands.
Loops
Secondary structures that are common include
scattered with areas of unsteady
structures that are called a
coil or a loop.
Typically, loop areas are found at the
protein's exterior.
These areas are capable of structural
importance and potential
the position of the useful part or
protein's "active site".
3D-Figure 3. PDB structure of 3H6R is a beta-trefoil cysteine protease inhibitor with sequence length of 152 (CLICK me for more info!!).
Ramachandran plot for all secondary structures has been depicted below.
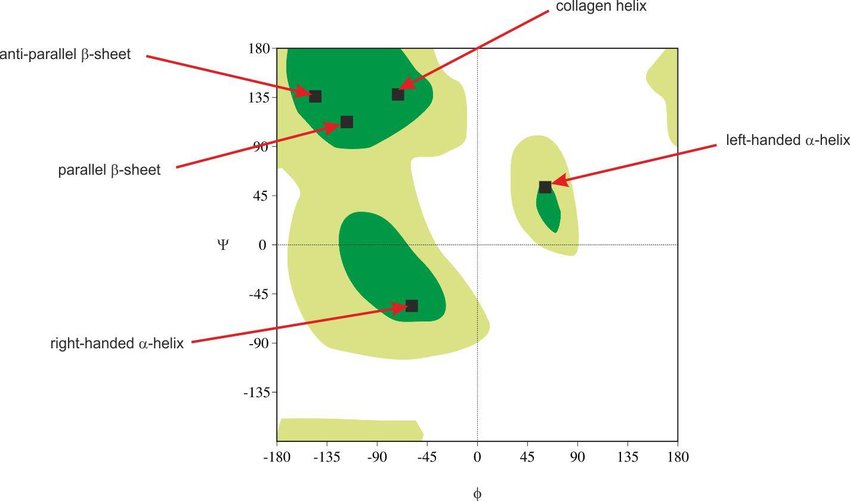
Figure 30. Ramachandran plot for all permissible regions for depicted supersecondary structures.
---- Summary ----
As of now you know all basics of Protein Structures.
-
Central Dogma of life.
-
Amino acids and it's types.
-
Peptide bonds and torsion angles.
-
Ramachandran Plots and protein secondary structures.
-
etc..
________________________________________________________________________________________________________________________________

________________________________________________________________________________________________________________________________
