4. Protein:
Parts of protein
Amino acids
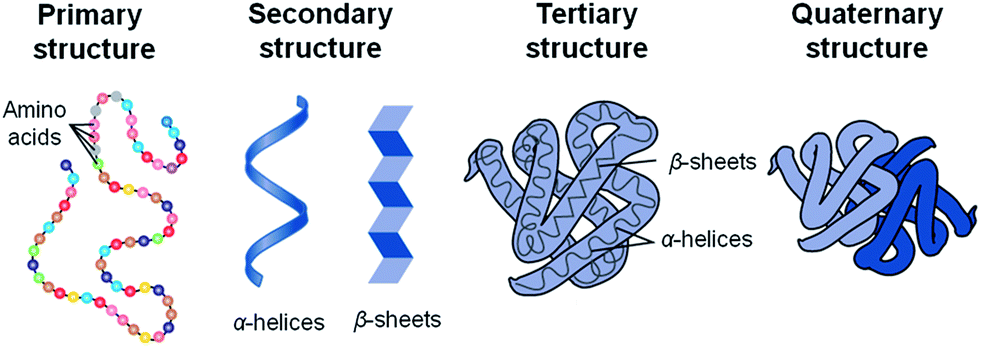
Figure 1. Protein organisational structures.
Peptide bonds connect the amino acids in a polypeptide chain to make proteins.
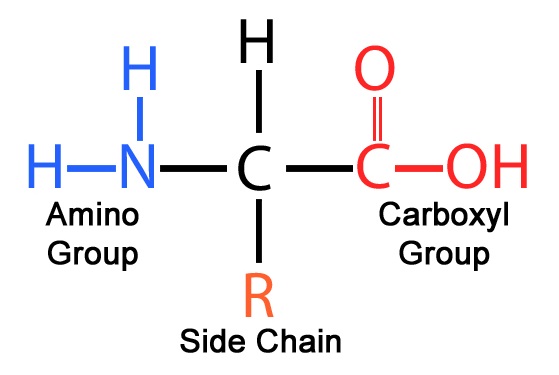
A variety of structural elements make up an amino acid:
Figure 2. A basic amino group (NH2), an acidic carboxyl group (COOH), and an organic R group (or side chain) that is specific to each amino acid make up an amino acid, an organic molecule.
A central carbon atom (Cα) is attached to :
-
An amino group (NH2)
-
A carboxyl group (COOH)
-
A hydrogen atom (H)
-
A side chain (R)
Polypeptide chain
Repeating units (residues) are divided into main chain (or backbone) and side chains (R1,R2,R3)
.png)
Figure 3. The internal angle of a polypeptide backbone where two adjacent planes meet is known as a protein's dihedral angle. Because the backbone located between two adjacent C alpha atoms is in a single plane, the conformation of the backbone can be represented by two dihedral angles per residue.
The main chain is identical in all residues, contains a central Cα atom attached to an NH group, a C'=O group, and an H atom.
Side chain, R, is different for different residues and bound to the Cα atom.
Disulphide bonds
The side chains of two cysteine residues generate disulfide bonds.
.png)
Figure 4. A disulfide is a functional group with the formula RSSR′ that is used in biochemistry. The connection, which is frequently created by the binding of two thiol groups, is also known as an SS-bond or occasionally as a disulfide bridge.
One S—S (disulfide) group is created by the oxidation of two SH groups from cysteine residues, which may be adjacent in the three-dimensional structure but in another location in the sequence.
.png)
Figure
5.
Disulfide bonds are covalent connections that develop
between the side chains of cysteines.
The chemical equation for creating a disulfide bond between two cysteine
side chains is shown in (A). Red is used to indicate the disulfide bond.
(B) A schematic of a disulfide bond (red) formed between two distinct
polypeptide chain sections is displayed (black). The polypeptide chain
is constrained into rigid conformations by the disulfide bonds, which
facilitates protein folding.
The side chain of the amino acid cysteine has a sulfhydryl (-SH) group. Sulfhydryls have the ability to be oxidized to create disulfide connections, which frequently involve two cysteine side chains from different positions in the main sequence.
Such disulfide link production in proteins makes the protein stiff and can stabilize conformations that are not generally preferred.
Cysteine is the collective name for a pair of cysteine residues that are disulfide-bonded.
Your hair serves as a well-known illustration of disulfide bonding. These connections are prevalent in hair because they are crucial to its strength. Beauty parlors benefit from your hair's disulfide connections.
.png)
Figure 6. the amino acids cysteine; cysteine amino acids can link with cysteines lower down the hair shaft because hair is primarily made of keratin, which contributes to the curling of the hair between the bonds.
If you were born with straight hair but now want curly hair, you can visit the salon and obtain a "perm." Your hairdresser treats your hair with a reducing agent, like thioacetic acid, which causes the disulfide bonds to dissolve and chemically change back into free cysteines.
3D-Figure 1. Pubchem structure of 10484 is of thioacetic acid with MW of 76.12 (CLICK me for more info!!).
In order to create new disulfide bonds, your hair is then treated with an oxidizing substance, typically hydrogen peroxide, after being curled around rollers to bring various sulfhydryl groups together. You now have curly hair instead of straight hair.
An organosulfur molecule called thioacetic acid has the molecular formula CH 3C(O)SH. The liquid is yellow and smells strongly of thiols.
Torsion angles in polypeptides
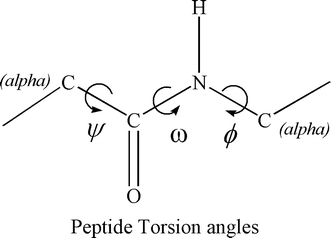
Figure 7. Phi (Ci-1 Ni Ca Ci) , Psi (Ni Ca Ci Ni+1) and omega (Ca Ci-1 Ni Ca)
phi (φ) is the C(i-1),N(i),Ca(i),C(i) torsion angle
psi (ψ) is the N(i),Ca(i),C(i),N(i+1) torsion angle.
The Phi and Psi angles determine the path of the polypeptide chain.
Alpha helices, beta sheets, and turns are three
frequently occurring and well-defined kinds of secondary structure in
proteins.
Loops are frequently referred to as regions other than clearly defined
secondary structure elements.
Combinations of specific phi and psi torsion angles and H bonds make up secondary structure.
Secondary structure elements
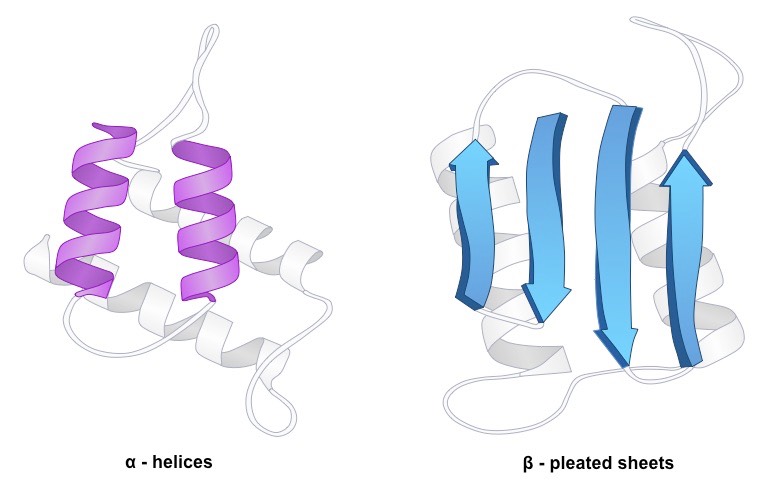
Figure 8. Secondary structure in a polypeptide chain refers to the predictable, recurrent spatial configurations of adjacent amino acid residues that are kept together by hydrogen bonds between the amide hydrogens and the carbonyl oxygens of the peptide backbone. α-helices and β-structures are the two main secondary structures.
Alpha chain
Main chain N and O atoms are hydrogen-bonded to one another; an alpha helix has 3.6 residues per turn, or 5.4 A. (rise of 1.5 A per residue)
Beta sheet
A typical motif of the typical protein secondary structure is the beta sheet, sometimes known as the "β sheet" or "β pleated sheet."
Beta sheets are made up of beta strands (-strands) that are joined laterally by two or three backbone hydrogen bonds, resulting in a sheet that is typically twisted and pleated.
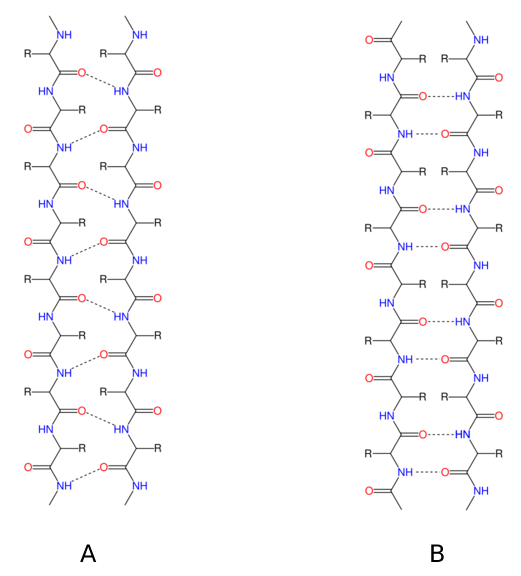
Figure
5. (A)
Patterns of parallel -sheet hydrogen bonds are shown as dotted lines.
Nitrogen atoms are shown blue, while oxygen atoms are red.
(B) Dotted lines depict antiparallel -sheet hydrogen bonding patterns.
Nitrogen atoms are shown blue, while oxygen atoms are red.
Twist of beta sheet
Although most sheets in globular proteins are
twisted (up to 30° per residue), theoretical beta sheets are planar.
Beta sheets that are anti-parallel are more frequently twisted than beta
sheets that are parallel, and two-stranded beta sheets exhibit the
biggest twists.
Different hydrogen bonding strengths contribute to twists.
3D-Figure 2. PDB structure of 1TI3 is a Thioredoxin h1 from poplar with sequence length of 113 (CLICK me for more info!!).
Loops
Loop backbone conformations are less well
defined than turn backbone conformations (more conformational freedom).
While loops are not specified secondary structures, turns are.
Loops often have a dense structural makeup with 6 to 16 residues.
Similar to twists, loops frequently have polar residues and are located
on the surface of proteins.
Secondary structure motifs
Configurations of a
small variety of secondary structure components that are found
repeatedly in protein complexes and have a particular geometric
arrangement.
Having a lot of structural or functional
importance.
Some illustrations of secondary structural themes are
:
-
Beta-Alpha-Beta
-
Helix-turn-Helix
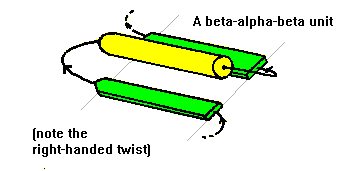
Figure 6. Two beta strands are connected by an alpha helix through connecting loops to form a beta-alpha-beta motif. The helix is nearly parallel to the beta strands and the strands are parallel to the beta strands. Almost all proteins with parallel strands display this structure.
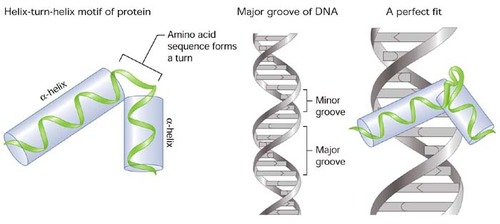
Figure 7. Proteins that bind DNA frequently contain the helix-turn-helix (HTH) motif, particularly transcription factors. The last helix of the HTH motif, also known as the recognition helix, inserts into the DNA major groove during DNA recognition to facilitate base-specific DNA recognition.
A double helix with an unpaired loop is created when two portions of the same strand, which are typically complementary in nucleotide sequence when read in opposing directions, base pair.
Many RNA secondary structures use the resultant structure as a fundamental building block.
Beta -Alpha -Beta
An alpha helix frequently connects two parallel
neighboring beta strands from strand 1's C terminus to strand 2's N
terminus.
The majority of parallel beta-sheet protein complexes are composed of a
mix of beta-alpha-beta motifs.
Helix-turn-Helix
A helix—turn—helix motif is made up of two alpha helices joined by a brief loop region in a particular geometric configuration:
(a) the DNA—binding motif
(b) the calcium- binding motif, which occur in many proteins.
Protein Domain
According to Richardson (1981), "contiguous
polypeptide chain segments typically fold into compact, local,
independent entities termed domains."
Each domain's fold can be used to characterize it because domains are
autonomous folding units.
Domains inside a protein can be thought of as interconnected units;
proteins can have one or many domains.
Domain Vs Motifs
Motifs do not function independently of domains.
Compared to motifs, domains are substantially larger.
A domain can be made up of many motifs.
Adjacent motif
Four Beta-Alpha-Beta-Alpha motifs that are consecutive in both the amino acid sequence and the three-dimensional structure make up triose-phosphate isomerase (TIM).
Tertiary structure
The term "tertiary structure" refers to the
three-dimensional association of secondary structure components or
domains inside a single polypeptide chain.
Three basic folding classes can be used to categorize tertiary
structures (each category may include numerous domains):
-
all Alpha folds
-
all Beta folds
-
Alpha/Beta folds
Alpha Folds Tetrad Helical Bundles
All-alpha proteins usually contain
four-helix bundles as domains.
In the center of the bundle, certain side chains from all four helices
are buried, creating a hydrophobic core.
Quaternary helix bundles
Contrary to human growth hormone, which has two pairs of parallel alpha helices, cytochrome b562 has neighboring helices that are anti-parallel.
The Globin folds
Globin folds
represent
a vast family of proteins that includes
hemoglobin and myoglobin.
In the globin fold, there are eight alpha helices (A-H).
Alpha and Beta folds
The most regular and prevalent domain structures
are made up of beta—alpha—beta motifs that repeat, resulting in an
outside layer of alpha helices packed against a core of parallel beta
sheets in the middle of the structure.
Alpha/beta proteins fold mostly into one of three different patterns:
-
Alpha//Beta (TIM) barrel fold
-
Open Alpha//Beta fold
-
Alpha/Beta horseshoe fold
TIM Barrel
beta strands that are tightly packed and twisted
at the center, surrounded by helices.
The consecutive connections between the beta-alpha-beta motifs create a
closed barrel shape.
The enzyme triose-phosphate isomerase has a prototype structure (TIM).
The alpha-beta barrel fold is a common one among enzymes.
Many different enzymes with entirely different
amino acid sequences and activities contain alpha/beta barrels.
The majority of these structures contain a closely packed hydrophobic
core made entirely of side chains from the barrel's eight beta strands.
Alpha-Beta Barrel Active site
The loop regions that link the C-terminal ends of the beta strands with the nearby alpha helices create a pocket that houses the active site in each barrel.
Catalytic sites
The barrel fold is a remarkably flexible
substrate for the development of various enzyme activity due to the
possibility of such small changes within an otherwise structurally
conserved scaffold.
Variations of residues inside the loops forming the pocket in Alpha/Beta
barrels can provide different active sites.
Open Alpha / Beta Fold
Alpha helices surround an open twisted beta sheet on both sides.
Examples of several open alpha/beta structure types include:
-
the enzyme adenylate kinase
-
the redox protein flavodoxin
Open Alpha/Beta folds active site.
On the opposing edges of the sheet, there are
two pairs of adjacent beta strands.
There is a gap at the edge of the beta sheets between the two loops
because one of the loops is above the sheet and the other below it.
The crevice contains almost all of the binding sites in this family of
proteins.
Beta Folds
Two beta sheets with anti-parallel beta strands typically pack against one another to create a sandwich-like shape. An example would be Superoxide dismutase (SOD)
Up & Down Barrel
Eight strands are arranged in an anti-parallel fashion and linked by beta hairpin loops.
A topological and schematic depiction of an up-and-down beta barrel.
Amino acid reflecting Beta structure
Aligned residues point into the barrel.
These mainly hydrophobic residues are denoted by green arrows. The
solvent comes into contact with the leftover residues.
Ile,Ala,Phe,Val,Met,Gly are examples of hydrophobic residues
Immunoglobin fold
Immunoglobulin's constant and variable domain beta strands, denoted A–G, share the same topology and structural similarities.
The variable domain has two additional strands,
C' and C" (red).
The hyper-variable region CDR2 is located in the loop that connects
these strands.
The remaining CDR regions are located in the loops that join strands B and C (CDR1) and F and G (CDR3) at the same end of the sandwich.
The lg fold
is the most significant module for mediating a range of various binding
interactions on the cell surface in addition to its function in creating
antibody combining sites.
For instance, extracellular lg domains are present on around 50% of
immunological cell surface receptors and ligands.
Homology and Similarity in Structure
Diverging proteins maintain their global fold during evolution.
Domains with homology have similar structures.
despite the modest sequence similarity.
1PLS/2DYN
23% Seq ID
Low sequence identity but some structure and function
The globin fold is not affected by variations in amino acids.
Although the eukaryotic hemoglobin F'. marinus and the bacterial hemoglobin V. stercoraria only have 8% sequence identity between them, their overall fold and function are the same.
Same Structure Different Functions
1ymv
1fla
1pdo
CheY
Flavodoxin
Mannose Transporter
Signal Transduction
electron transport
Less than 15% sequence identity
Local sequence similarity does not equate to structural or functional similarity on a global scale.
viral capsid protein glycosyltransferase
80 residue stretch (yellow), with a sequence identity of more than 40%
Convergent evolution
Both chymotrypsin and subtilisin are serine
proteases.
Their folds are unconnected, and they have no shared sequence identity.
However, they share a Ser- His-Asp catalytic triad that is conserved in three dimensions, which catalyzes the hydrolysis of peptide bonds.
-
Subtilisin EC 3.4.21.62
-
Chymotrypsin EC 3.4.21.1
Superfolds
Superfolds are classified as structure
with One-domain superfolds being common.
Despite protein divergence during evolution, global folds are preserved.
Protein structure is substantially more conserved than protein sequence
during evolution.
Superfolds are protein folds that occur frequently despite minimal
sequence similarity or the absence of evolutionary links.
2IMM Immunoglobulin like
ITPH Beta/alpha barrel
1UBI Beta grasp
1CKA SH3 Like barrel
3CHY Flavodoxin
1FXD Ferredoxin
1SNC OB-fold
1NFN 4-Helix Bundle
Structural Classification: SCOP
Protein classification based on structure.
All proteins with known structures are arranged in the SCOP database
according to their structural and evolutionary links.
automated techniques supporting manual comparisons and visual assessment
(structural competence).
four hierarchical categories make up this:
-
Class: types of folds, e.g. beta sheets.
-
Fold : varying shapes of domains inside a class.
-
Superfamily: grouping domains in a fold into superfamilies that share at least a remote ancestor.
-
Family: superfamily domains are organized into families that share a more recent common ancestor.
Structure-Based Semi-Automated Domain Definition: CATH
CATH hierarchy:
-
Class
-
Architecture
-
Topology
-
Homology
Classes involves automated detection of secondary structure content.
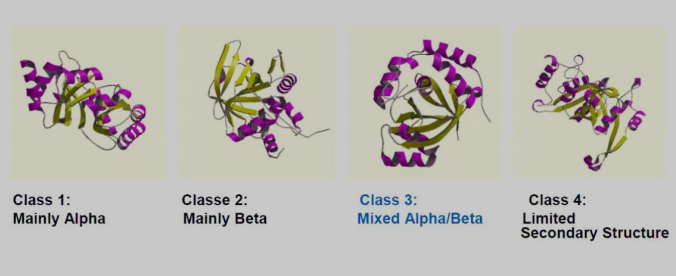
-
Class 1: Mainly Alpha
-
Class 2: Mainly Beta
-
Class 3: Mixed Alpha/Beta
-
Class 4: Limited Secondary Structure
Architecture involves manual fold assignment and relative alignment of secondary structural parts.
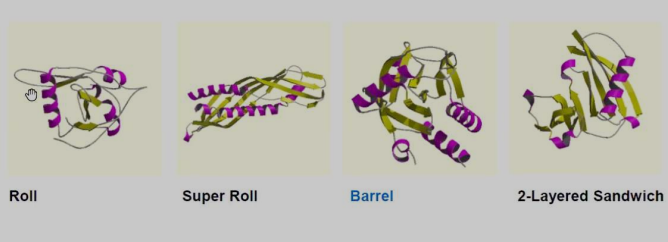
-
Roll
-
Super Roll
-
Barrel
-
2-Layered Sandwich
Topology includes automatic determination of number of secondary structures and topological relationships.
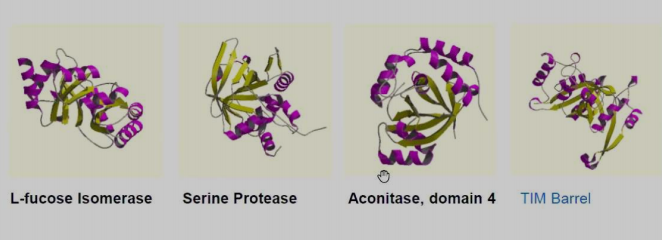
-
L-fucose lsomerase
-
Serine Protease
-
Aconitase, domain 4
-
TIM Barrel
Homology has superfamilies, which are collections of related structures and functions, follow both manual and automatic management techniques.
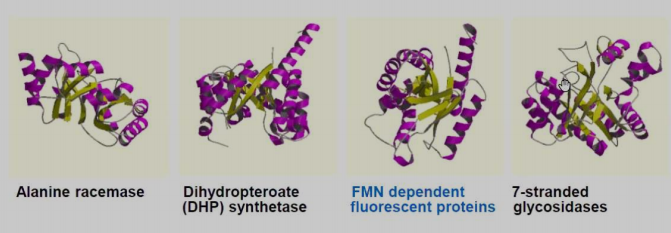
-
Alanine racemase
-
Dihydropteroate (DHP) synthetase
-
FMN dependent fluorescent proteins
-
7-stranded glycosidases
---- Summary ----
As of now you know all basics of Fold Structures.
-
Domains and motif.
-
Various secondary structure motifs.
-
SCOP.
-
CATH.
-
etc..
________________________________________________________________________________________________________________________________

________________________________________________________________________________________________________________________________